7. Codes
7.1. IMASviz
The IMASViz code is used for IMAS visualisation.
7.2. IMASgo!
IMASgo! is an OMFIT module for the mapping of experimental data and machine descriptions into an IMAS database. IMASgo! builds upon the OMFITprofiles, KineticEFIT and TRANSP modules. Any tokamak for which the above modules have been configured can use IMASgo! to map its data into IMAS. At present IMASgo! can write IDSs on the ITER server and on the WPCD Gateway. An account on one of these servers is necessary to complete the workflow in IMASgo! IMASgo! workflow starts with reading the equilibrium from the Tokamak database; kinetic measurements such as Thomson Scattering, ECE, Charge Exchange will be then mapped on the equilibrium and fitted with a variety of available methods. The last step is to export the experimental data, the fitted profiles and the machine description of NBI and RF antennas in the IMAS database. Below are the step by step instructions on how to launch the IMASgo! module and perform the mapping for JET. Some of the files used by IMASgo! for the mapping of the NBI / ICRH machine data are only accessible from inside the JET network hence this example has been run on Heimdall
On an Heimdall terminal type
$ module purge
$ module load omfit
$ omfit
The OMFIT framework will be launched Click on continue to OMFIT. From the File drop menu select
Select the module from the list of available modules.
Double click on IMASgo. The module will be loaded in OMFIT and ready to be launched.
Double click on IMASgo in the View1 list of loaded modules.
A GUI will be displayed. In this case the device choses is JET and the pulse and Times to be mapped appear in the next two fields at the top of the GUI.
From the drop menu Operation chose equilibrium from PPF It is possible to choose amongst different EFIT++ option: run EFIT++, load the chain 1 magnetics only EFIT, load the pressure constraint equilibrium in the drop menu and selecting the PPF UID and sequence number.
Click on Generate equilibrium.
Once the step is completed click on the tab Profiles and select 1D fits from the drop menu .
Click the Fetch tab and select which diagnostics you would like to upload data from.
Click on the fetch and map all data buttons.
In the Slice tab select the time averaging and click slice all data.
In the select tab you can visualise the sliced profiles and deselect profiles in advance of the the fitting step. Once the selected profiles are ok move to the fit tab and select the fitting method for all the data that need fitting. Chose the default fitting parameters or modify them. Then click on fit 1D and plot.
Once all the data are fitted move to the postfit tab and calculate the derived quantities.
The postfit tab allows also for manipulation of the profile in order to meet certain constraints e.g. separatrix values. The plot tab allows to have a final look at the data before they are saved in the database.
Click on the Machine tab and click on Generate Machine Description. This step will provide the data for the NBI and ICRH IDSs.
The final step is to export the data.
Click on the Export tab and on Generate OMAS. This will save the data in memory into the OMAS datastructure.
Then set up the server, shot number, run number and hit save ODS to IMAS. An entry will be created in the user’s IMAS database on the Gateway for JET/92436/107 as well as the same entry for the ITM database (CPOs).
Two videos showing an example of use of IMASgo! to fetch and map data of JET pulse 92054 (NBI only) and run ETS with the same data are available on YouTube at
7.3. How to turn a C++ code into a Kepler actor
This document is based on material provided by Yann Frauel and describes how to make your C++ code EU-IM compliant and how to turn it into a Kepler actor.
7.3.1. Adapt your C++ function
You must include the header file UALClasses.h:
#include "UALClasses.h"
The function arguments that are arrays or strings must be declared as pointers, as usual. All other arguments must be passed by reference (i.e. they must be declared with an ampersand):
void mycppfunction(double * vector, char * string, int & scalar)
The function arguments that are CPOs must be declared with types ItmNs::Itm::cpo_type or ItmNs::Itm::cpo_typeArray. The first form is for time-independent CPOs or a single slice of a time-dependent CPO. The latter is for a complete time-dependent CPO. Note that in all cases, the CPO is considered as a single object, not an array, so it must be passed by reference as mentioned above:
void mycppfunction(
ItmNs::Itm::limiter & lim,
ItmNs::Itm::coreimpur & cor,
ItmNs::Itm::ironmodelArray & iron)
The syntax is identical for input and output arguments. For output CPOs, do not forget to use the usual methods to assign strings and allocate arrays:
lim.datainfo.dataprovider.assign("test_limiter");
iron.array.resize(3);
iron.array(j).desc_iron.geom_iron.npoints.resize(3);
Otherwise, the content of CPOs is accessed as usual:
cout << lim.datainfo.dataprovider << endl;
cout << iron.array(j).desc_iron.geom_iron.npoints(i);
7.3.2. How to use code parameters
The code parameters are passed as the last argument with ItmNs::codeparam_t& type:
void mycppfunction(..., ItmNs::codeparam_t & codeparam)
Each field of the param structure is a vector of 132-byte strings, not necessarily terminated by 0-character! (This does not follow C/C++ standards and should be changed in the future.)
7.3.3. Compile your function as a library
You need to include the header directories for the UAL and Blitz:
-I$(UAL)/include -I$(UAL)/lowlevel -I$(UAL)/cppinterface/ -I/afs/efda-
itm.eu/gf/project/switm/blitz/blitz-0.9/include/
Same for linking:
-L$(UAL)/lib -lUALCPPInterface -lUALLowLevel -L/afs/efda-
itm.eu/gf/project/switm/blitz/blitz-0.9/lib -lblitz
Additionally, you must compile with the -fPIC option.
7.3.4. Full example
We want to generate an actor that has three different types of actors as inputs and three different types of actors as output. Additionally, we have an integer as input/output, a vector of doubles as output and a string as output. We also want to use code parameters. Content of mycppfunction.cpp:
#include "UALClasses.h"
typedef struct {
char **parameters;
char **default_param;
char **schema;
} param;
void mycppfunction(
ItmNs::Itm::summary SUM,
EU-IMNS::EU-IM::ANTENNAS & ANT,
EU-IMNS::EU-IM::EQUILIBRIUMARRAY & EQ,
INT & X,
EU-IMNS::EU-IM::LIMITER & LIM,
EU-IMNS::EU-IM::COREIMPUR & COR,
EU-IMNS::EU-IM::IRONMODELARRAY & IRON,
DOUBLE * Y,
CHAR * STR,
PARAM & CODEPARAM)
{
/* DISPLAY FIRST LINE OF PARAMETERS */
COUT << codeparam.parameters[0] << endl;
cout << codeparam.default_param[0] << endl;
cout << codeparam.schema[0] << endl;
/* display content of inputs */
cout << "x=" << x << endl;
cout << sum.time << endl;
cout << sum.datainfo.dataprovider << endl;
cout << ant.datainfo.dataprovider << endl;
cout << eq.array(0).datainfo.dataprovider << endl;
for (int k=0; k<3; k++) {
for (int i=0; i<4; i++) {
cout << eq.array(k).profiles_1d.psi(i)<< " ";
}
cout << endl;
}
/* fill limiter CPO */
lim.datainfo.dataprovider.assign("test_limiter");
lim.position.r.resize(5); // allocate vector
for (int i=0; i<5; i++) {
lim.position.r(i)=(i+1);
}
/* fill coreimpur CPO */
cor.datainfo.dataprovider.assign("test_coreimpur");
cor.flag.resize(3); // allocate vector
for (int i=0; i<3; i++) {
cor.flag(i)=(i+1)*10;
}
cor.time=0; // don't forget to fill time for time-dependent CPOs
/* fill ironmodel CPO */
iron.array.resize(3); // allocate slices
for (int j=0; j<3; j++) {
char s[255];
sprintf(s,"test_ironmodel%d",j);
iron.array(j).datainfo.dataprovider.assign(s); // allocate vector
iron.array(j).desc_iron.geom_iron.npoints.resize(3);
for (int i=0; i<3; i++) {
iron.array(j).desc_iron.geom_iron.npoints(i)=j*i;
}
iron.array(j).time=j; // fill time for time-dependent CPOs
}
/* assign value to non CPO outputs */
x=5;
for (int i=0; i<10; i++) {
y[i]=i;
}
strcpy(str,"This is a test string");
}
Content of Makefile:
CXXFLAGS=-g -fPIC -I$(UAL)/include -I$(UAL)/lowlevel -I$(UAL)/cppinterface/
-I$SWEU-IMDIR/blitz/blitz-0.9/include/
LDFLAGS=-L$(UAL)/lib -lUALCPPInterface -lUALLowLevel -L/afs/efda-
itm.eu/gf/project/switm/blitz/blitz-0.9/lib -lblitz
libmycppfunction.a: mycppfunction.o
ar -rvs libmycppfunction.a mycppfunction.o
mycppfunction.o: mycppfunction.cpp
clean:
rm mycppfunction.o libmycppfunction.a
7.3.5. How to fill the FC2K window
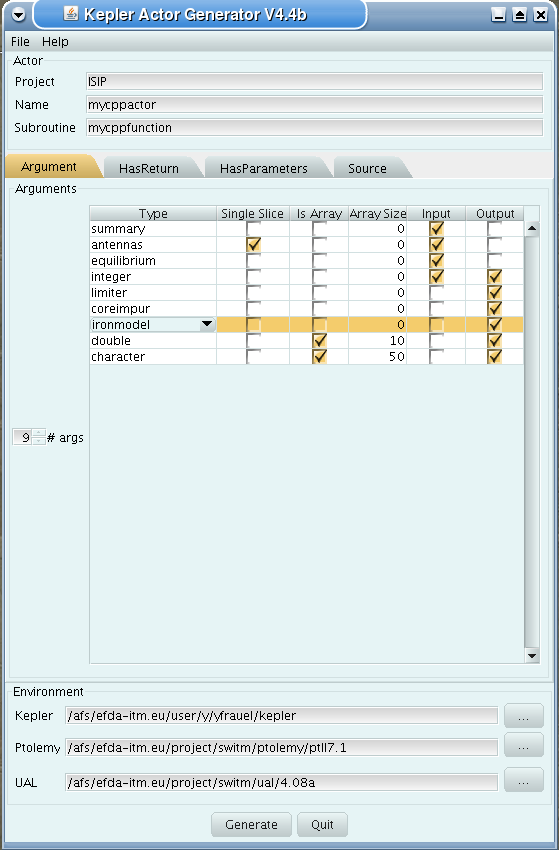
First tab (Argument):
set number of input and output arguments (combined)
select type of arguments from drop-down menu
tick if argument is a single time slice
tick if argument is array (not for pointers)
if necessary define size of arrays
tick if argument is input argument
tick if argument is output argument (multiple ticks possible)
The fields Kepler, Ptolemy, and UAL are automatically filled with the
values which you set by running the EU-IMv1 script.
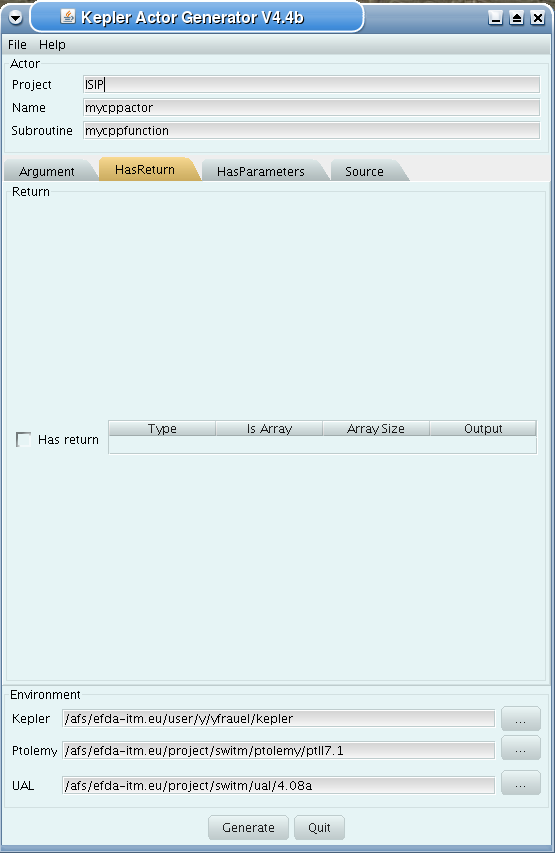
Second tab (HasReturn):
specify return parameters (type, array, size)
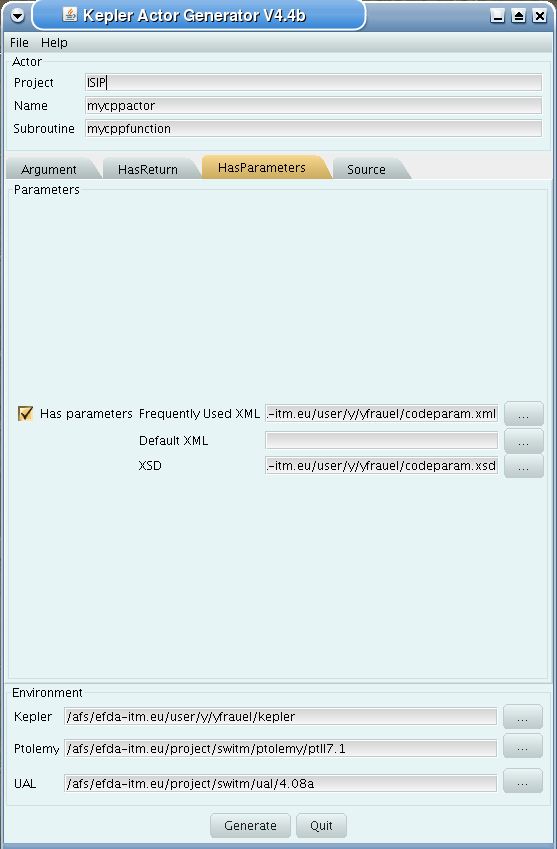
Third tab (HasParameters):
tick if subroutine uses code specific parameters
specify (or browse for) XML code parameter input file
specify (or browse for) XML default code parameter file
specify (or browse for) W3C XML schema file (XSD)
For information on code specific parameters, please see How to handle code specific parameters.
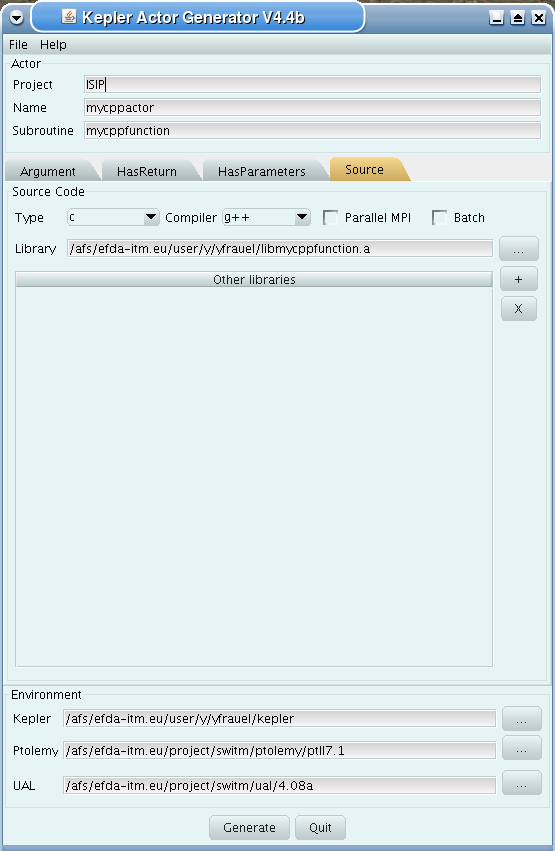
Fourth tab (Source):
specify programming language of source code
select appropriate compiler
tick Parallel MPI if code module is using MPI
tick Batch if code module shall be run in batch mode rather than interactively when running Kepler workflows
specify (or browse for) library file containing the code module
specify (or browse for) other libraries required by the code module
7.4. Plasma equilibrium and MHD list of codes
The following list lists the codes and modules which are part of WPCD tasks and their responsible officers.
7.4.1. Free boundary equilibrium codes
CEDRES++, S. Brémond, CEA
CLISTE, P. Mc Carthy, DCU
CREATE-NL, M. Mattei, ENEA Frascati
EFIT++, L. Appel, CCFE
EQUAL, W. Zwingmann, EC
EQUINOX, B. Faugeras, CEA
FIXFREE, E. Giovannozzi, ENEA Frascati
7.4.2. Fixed boundary equilibrium codes
CAXE, S. Medvedev, EPFL
CHEASE, O. Sauter, EPFL
HELENA, C. Konz, IPP
7.4.3. Linear MHD stability codes
KINX, S. Medvedev, EPFL
ILSA, C. Konz, IPP
MARS, G. Vlad, ENEA Frascati
MARS-F, D. Yadykin, Chalmers
7.4.4. Sawtooth Crash Modules
SAWTEETH, O. Sauter, CRPP
7.4.5. ELM Modules
7.4.6. NTM Modules
NTMETS, S. Nowak
7.4.7. Numerical Tools
PROGEN, C. Konz, IPP
JALPHA, C. Konz, IPP
7.5. Heating, current drive (H&CD) and fast particles list of codes
The following list lists the codes and modules which are part of WPCD tasks and their responsible officers.
7.5.1. Electron heating codes
7.5.1.1. EC wave codes
TORAY-FOM, E. Westerhof, FOM
TORBEAM, E. Poli, IPP-Garching
GRAY, L. Figini, ENEA-CNR
TRAVIS, N. B. Marushchenko, IPP-Greifswald
7.5.1.2. Combined electron Fokker-Planck codes
RELAX, E. Westerhof, FOM
7.5.1.3. Wave codes for ion cyclotron heating
TORIC, R. Bilato, IPP-Garching
EVE, R. Dumont, CEA (Cadarache)
LION, O. Sauter, CRPP
Cyrano, E. Lerche, ERM/KMS
ICCOUP, T. Johnson, VR
7.5.1.4. Fokker-Planck codes for ion cyclotron heating
RFOF, T. Johnson, VR
StixRedist, E. Lerche and D. Van Eester
7.5.1.5. NBI sources for Fokker-Planck codes
BBNBI (Beamlet-based NBI module of ASCOT), J. Varje, TEKES
NEMO, M. Schneider, CEA (Cadarache)
7.5.1.6. Nuclear sources (input for Fokker-Planck codes)
Nuclearsim, T.Johnson, VR
AFSI Ascot Fusion Source Integrator, J. Varje, Aalto
7.5.1.7. NBI Fokker-Planck codes
RISK, M. Schneider, CEA (ITER)
NBISIM, T. Johnson, VR
7.5.1.8. Runaway electrons
Runaway Indicator (Runin), G. Pokol, et al (BME): Runin has been developed to provide an indication for when to expect runaway tail formation. The source code is stored in the OSREP project <https://github.com/osrep>.
Runaway Fluid (Runafluid), G. Pokol, et al (BME): Porpose of Runafluid is to provide a non-inductive current due to runaway electrons using computationaly cheap analytical estimates of ruraway electron growth rates and transport. The source code is stored in the OSREP project <https://github.com/osrep>.
7.5.1.9. Advanced codes
(The following codes include either the synergy between IC and NBI heating, or include both wave field and Fokker-Planck solver)
ASCOT, S. Sipila and J. Varje, Aalto
SPOT, M. Schneider, CEA (Cadarache)
7.5.1.10. Codes for fast ion-MHD interactions
LIGKA, P. Lauber, IPP-Garching
MARS, G. Vlad, ENEA-Frascati
HYMAGYC, G. Vlad, ENEA-Frascati
HMGC, C. Di Troia, ENEA-Frascati
LEMAN, W.A. Cooper, EPFL-CRPP
7.6. Transport list of codes
ASPOEL
BIT1
CARRE
COS
EIRENE
EIRENE2
EMC3-EIRENE
ERO
ETS
METIS4EU-IM
SOLPS
SOLPS6